The central Dogma of Biology posits that information stored in DNA nucleotide triplets is transcribed into RNA which is then translated into a chain of amino acids .
Currently there are 2 opposing theories for the origin of the genetic code :
1. The Code was initially made from GC, then GCA and finally from GCAT resulting in the first 4 amino acid peptides consisting of Glycine, Alanine, Proline, and Diamino proprionic acid.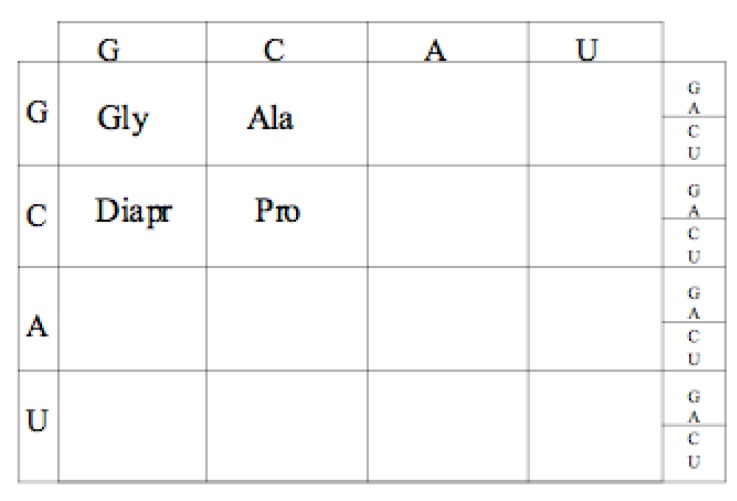
2. The Code was initially made from GNC encoding the amino acids of Glycine, Alanine, Valine, Aspartic Acid. 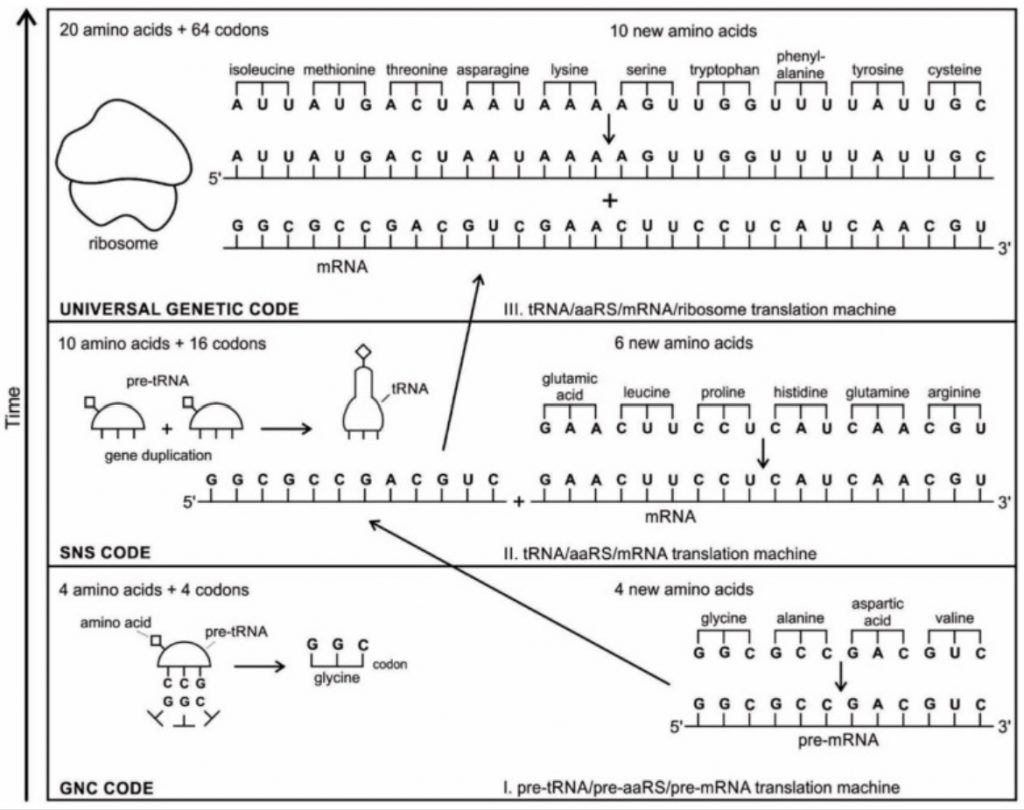
Both theories state that the code subsequently expanded in stages to include the full alphabet of amino acids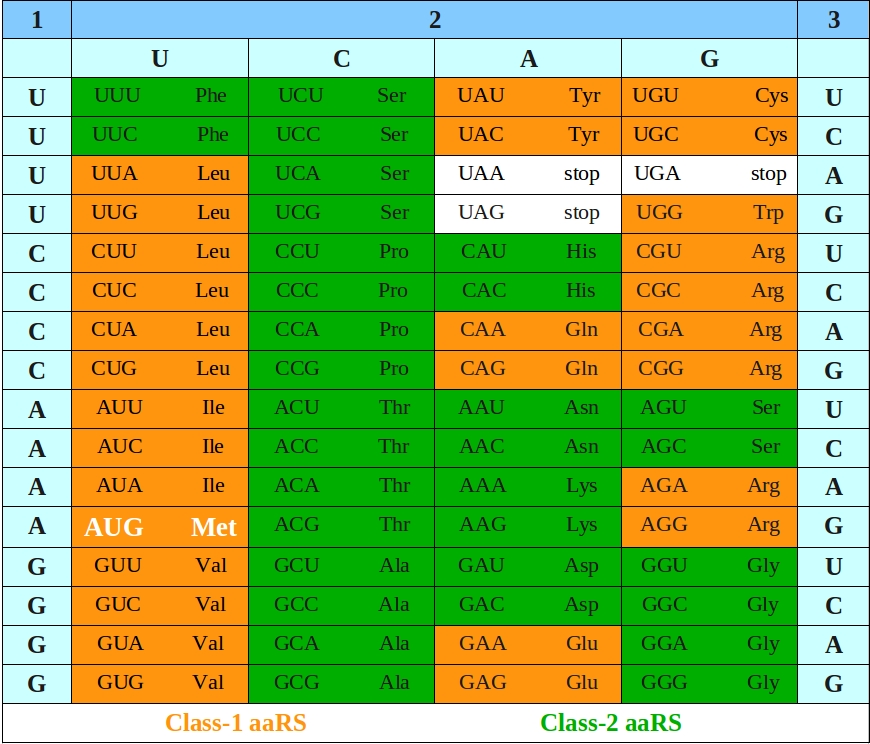
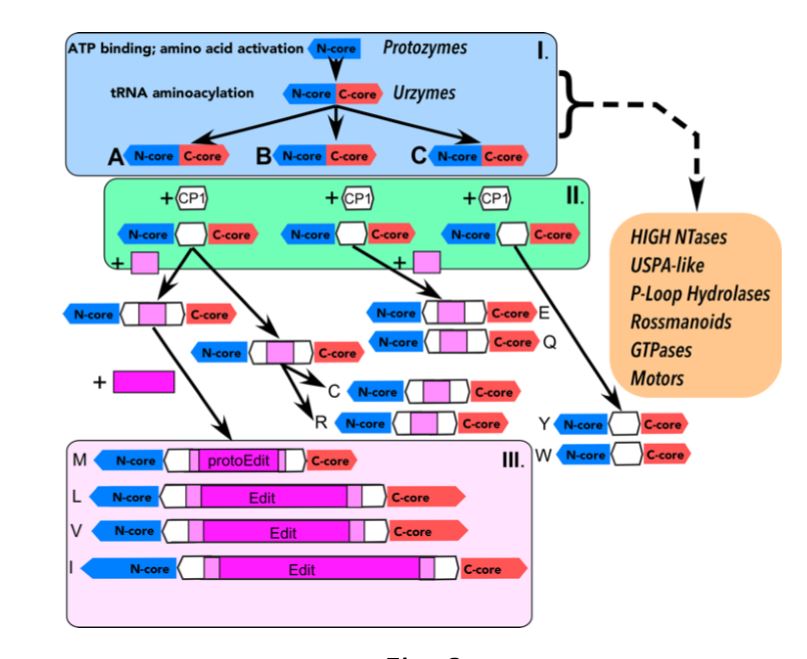
The first encoded proteins were amino acid synthase type I and II urzymes and protozymes that could be transcribed from the opposite strands of a double stranded 21 nucleotide helical RNA herein named urRNA (a precursor to siRNA, microRNA, PIWI-RNA, endoriRNA, tRNA, mRNA, rRNA, and DNA) .
The protozymes evolved into multiple protein super fold families such as the Rossman, OB, SH3, and TIM folds. The evolution of these protein fold super families occurred in stages proposed below:
Pre 1 Stage : Peptides and Microhelixes ligated to form proteins , tRNA, rRNA, and mRNA. The proteins evolved secondary structure that followed the pattern of (1) loops with beta turns; (2) beta hairpins; (3) alpha beta proteins; and then (4) alpha proteins
Stage 1. Proteinocytes/ Proteoids were the first life forms to replicate through non ribosome protein synthesis. They encapsulized the coacervates/ proteinoid microspheres with pseudomurein and contained the Citric Acid Metabolic Cycle of metabolites.
Stage 2. Inteinocytes are PreRNA based Proteinocytes and replicate through non ribosome protein synthesis. Inteinocytes contain self and non self splicing inteins. They have not yet evolved messenger RNAs but have Ribozymes, Riboswitches, Group I and II Introns, pretRNA/Ribosomal RNAs, and other non coding RNAs.
Stage 3. Transpocytes are RNA based proteinocytes. They have a peptidyl transfer system that utilizes RNA ligation and polymerization enzymes, polyketide synthesis, nonribosomal peptide synthetases, and ubiquitin conjugation systems .. The ribosomal system is still evolving along with RNA replication/transcription systems.
RNA transpocytes are inteinocytes with replicative RNA
DNA transpocytes are inteinocytes with replicative RNA and DNA
Stage 4. Chronocytes/Virocytes are transpocytes with lipid membranes and transitional RNA/DNA genetic storage systems. They Operate with specific operons for hyperthermophilic, mesophilic, and psychrophilic environments. Their genomes are primarly RNA based.
Stage 5. Eocytes ( including LUCA -the last universal common ancestor ) containing DNA and multiple types of membranes gave rise to Prokaryotes and subsequently Eukaryocytes. They were delivered to Earth during the Theia impact and the late heavy bombardment. They are probably present on other solar system bodies such as CERES and Mars. Their origin can be traced to the solar systems Proplyd membrane stage of Solar System Evolution.
Possible ribosomal RNA CREATION Scenario : Posits early tRNA concatenated/ligated into self replicating ribosomal RNA + code for ribosomal proteins, transcription, replication, translation and metabolic proteins
Evolution of the ribosome in phases 1-8. Phase 1-4 represent pre-LUCA time periods.
Phase 1: Folding, rudimentary binding, and catalysis. AESs 1–2 is
a branched duplex with a defect that forms the P loop. This defect may confer catalytic activity and/or ability to bind specifically to small molecules.
Phase 2: Maturation of the PTC and formation of an exit pore.
Inclusion of AESs 3–5 adds the A region to the P region, in concert with formation of an exit pore.
Phase 3: Early tunnel extension. Inclusion of AESs 6–10 extends
the exit pore, creating a short tunnel. The stability and rigidity of the tunnel are increased by buttressing.
Phase 4: Acquisition of the SSU interface. AESs 11–28 are included. AESs 11, 12, and 15 form the LSU interface for association with the SSU. The other segments enhance the stability and efficiency of the LSU by embracing the
PTC and further extending the exit tunnel.
Phase 5: Acquisition of translocation function. Inclusion of AESs
29–39 adds essential components of the modern energydriven translational machinery: the L7/L12 stalk and central protuberance , and binding site (sarcin–ricin loop) for elongation factors G and Tu .The tunnel is further extended.
Phase 6: Late tunnel extension. Further expansion of the LSU by
inclusion of AESs 40–59 results in the maturation of
common core of the LSU. In the final phase of prokaryotic ribosomal evolution , the exit tunnel is extended. A majority of
elements added here are located at the ribosomal surface and interact with ribosomal proteins.
Phase 7: Encasing the common core (simple eukaryotes). Eukaryotic expansion segments are acquired and previous
AESs are elongated. This eukaryotic-specific rRNA
combines with eukaryotic-specific proteins to form a shell around
the common core.
Phase 8: Surface elaboration (complex eukaryotes). Metazoan
ribosomes are decorated with “tentacle-like” rRNA elements that extend well beyond the subunit surfaces. These tentacles are fundamentally different in structure and function than common core rRNA. Metazoan expansions appear.
The ribosome as a missing link in the evolution of life
Scenario 1 : Selfish Ribosomal RNA evolved first and produced 20 tRNAs + ribosomal protein, replication, and metabolic protein mRNA.
Scenario 2 : early tRNA concatenated/ligated into self replicating ribosomal RNA + code for ribosomal proteins, transcription, replication, translation and metabolic proteins .
Structural Phylogenomics Retrodicts the Origin of the Genetic Code and Uncovers the Evolutionary Impact of Protein Flexibility
Evolution of the ribosome and the genetic code
” It is in the proteins of the ribosome that the “fossil” record of the evolution of the Genetic Code is most apparent. Clearly the expansion of the Code involved the expansion of the amino acid carriers, tRNAs, also that of the mRNA system and finally that of the modern tRNA synthetases. The latter in particular required multiple domain protein complexes to be able to read both the assumed older information in the acceptor stem and in the anti-codon loop. Such complex proteins required the expanded code coupling the evolution of protein structure to that of the code”