Cellular
life forms are like Matryoshka/Babushka Dolls that show the history
of our earliest ancestors in the center and novel life forms in the
periphery. The history of the conversion of free living life forms
to free living parasites, symbionts, and agents of genetic change
shows a helical evolutionary pattern. The mobile genetic elements
show a Mirror in Mirror display of newer genetic elements nesting
in the center of older genetic elements . This nesting phenomenon
is exemplified by insertion
sequences in archaea and the insertion
history of DNA transposons. The encapsulation processes work
in tandem with the integration processes to evolve life. Analysis
of twintrons, symbioses, and other forms of nesting will help unravel
the amazing pathways of evolution.
What
cellular life form developed first on Earth?
1.
Archaea/Eocytes arose before Eubacteria
Support for this scenario can be seen in the evolution of the Nif
H gene in archaeic bacteria followed by monodermal and then didermal
eubacteria. See (Expression
and Association of Group IV Nitrogenase NifD and NifH Homologs in
the Non-Nitrogen-Fixing Archaeon Methanocaldococcus jannaschii and Origin
and Spread of Bacteriochlorophyll Biosynthesis Proteins. )
"The
evolutionary history of protein fold families and proteomes confirms
that the archaeal ancestor is more ancient than the ancestors of
other superkingdoms . "
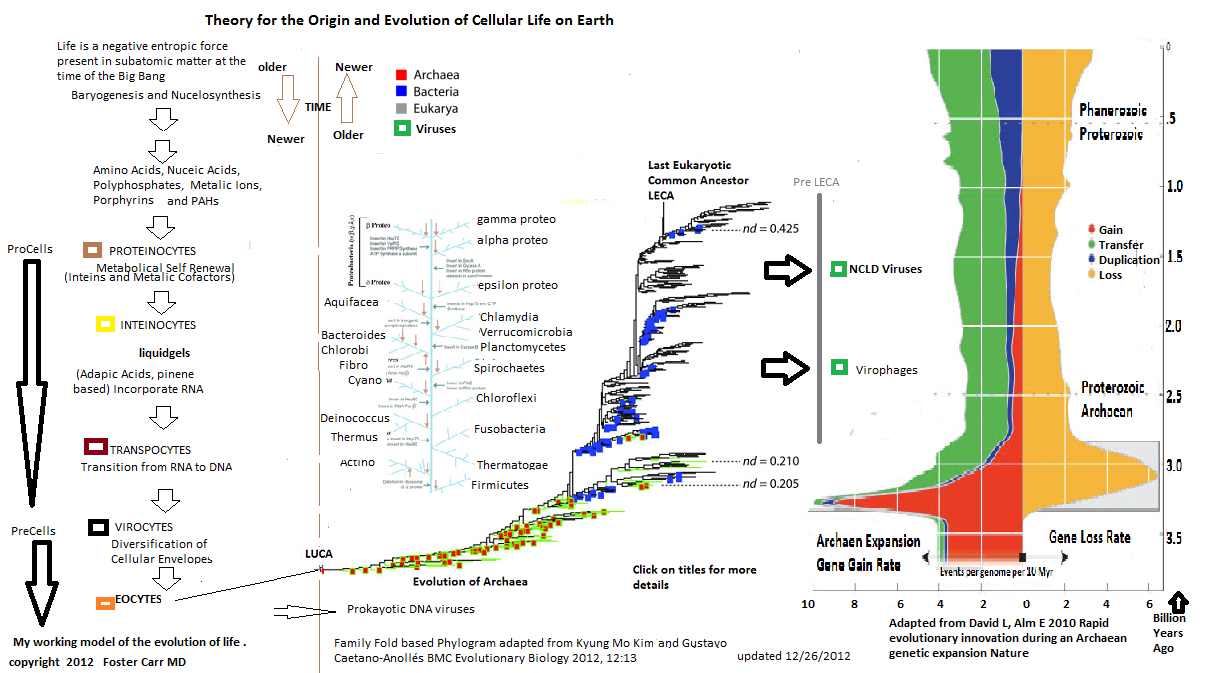
2. Archaea and Eubacteria
evolved separately and at the same time.
Forterre proposes that viral infection
events created 3 separate superkingdoms and thus there is no
monophyly of all cellular life forms. (see Three
RNA cells for ribosomal lineages and three DNA viruses to replicate
their genomes: a hypothesis for the origin of cellular domain)
3. Archaea are derived from
Eubacteria .
Cavalier
Smith proposes that Chlorobacteria form the root of the
prokaryote tree and archaea are derived from Actinobacteria.
Lake proposes
a circular tree root between firmicutes and actinobacterial ancestors.
He suggests that archaea are derived from the Firmicutes.
Valas
and Bourne propose that Archaea are derived from Neomura
which arose from Firmicutes/Clostridia due to a DNA virus infection
A refined understanding
of origin requires separating the phylogenetic history of transcriptional,
translational, replicative, metabolic. and
structural genes of early life forms. The answer to the question "Which
genes arose first ?" provides better insights into the evolution
of cells within the framework of the Public
Goods Hypothesis.
Forterre suggests that eubacteria,
archaea, and eukaryotic DNA replication arose from independent
viral infection of pre-archaeal ribosome containing cells (chronocytes).
Similarly, my working model suggests that Eocytes developed the
first replicative metabolic systems with stable membranes. These
Eocytes
evolved
into archaea and were later transformed by viral infections and
other symbiotic processes into the Eubacterial and Eukayotic lineages.
I speculate that symbiotic forms of Eocytes and Pre LECA cells
evolved into bacteriophages, virophages, and NCLD viruses. (
see 4th
NCLDV viral domain of dna replication proteins rooted between
the archaea and eukaryota and Phylogenetic
and Phyletic Studies of Informational Genes in Genomes Highlight
Existence of a 4th Domain of Life Including Giant Viruses)
Did monoderm prokaryotes evolve
into diderms or vice versa ?
Current data
supports a model of monoderms evolving into non-LPS diderms followed
by diderms with
LPS. see
(Origin
of diderm (Gram-negative) bacteria: antibiotic selection pressure
rather than endosymbiosis likely led to the evolution of bacterial
cells with two membranes ) There is also evidence that a branch
of monodermic actinobacteria independently evolved into didermal
corynebacteria with mycolic acids in their outer membrane. Furthermore,
the Negativicutes order from gram positive clostridia related
firmicutes appears to have independently evolved a didermal membrane.
Another possibility is that anoxygenic photosynthetic
halobacteria sporolated in the presence of
a diderm promoting plasmid. The resulting proto endospore became
the first diderm during the Hadean Epoch. The primordial diderm
was
a LPS negative, didermal deinococcus-thermus
organism. These hadobacteria then evolved into LPS positive cyanobacteria
as proposed by Cavalier Smith. Multiple horizontal gene transfers
where involved in the transition from anoxygenic monodermal photosynthesis
to didermal oxygenic photosynthesis during the Archaen
Epoch.
Alternatively, photosensitive nanohaloarchaea
evolved into photosynthesizing heliobacteria followed by an endosymbiosis
with actinobacteria to create cyanobacteria. Similar archaea +
gram positive
eubacterial
symbioses/lateral gene transfers or gram positive + gram positive
symbioses gave rise to the separate gram negative bacteria families.
(such
as
proteobacteria,
spirochaetes, FCB group, and PVC super group )
How did mobile
genetic elements effect the origin and evolution of prokaryotes?
How did the physiochemistry
of our Solar System, atmosphere ,
hydrosphere, and crust evolve life ?
What
is the origin and evolution of prokaryotic vesicles ?
Phylogenetic
Trees
, Species
Trees, Taxonomy
Browser of Euryarchaea and Eubacteria
News:
Proc Natl Acad
Sci U S A. 2012 Dec 11;109(50):20537-42.
Epub 2012 Nov 26.
Acquisition of 1,000 eubacterial genes physiologically transformed
a methanogen at the origin of Haloarchaea.
Nelson-Sathi S, Dagan T, Landan G, Janssen A, Steel M, McInerney JO,
Deppenmeier U, Martin WF. Narasingarao, Priya , Podell, Sheila, Ugalde, Juan A, Brochier-Armanet,
Celine, Emerson, Joanne B, Brocks, Jochen J , Heidelberg, Karla B ,
Banfield, Jillian F , Allen, Eric E
De novo metagenomic assembly reveals abundant novel major lineage
of Archaea in hypersaline microbial communities International Society
for Microbial Ecology ISME J 2012/01
Rohit
Ghai, Lejla Pašic, Ana Beatriz Fernández, Ana-Belen
Martin-Cuadrado, Carolina Megumi Mizuno, Katherine D. McMahon, R. Thane
Papke, Ramunas Stepanauskas, Beltran Rodriguez-Brito, Forest Rohwer,
Cristina Sánchez-Porro, Antonio Ventosa & Francisco Rodríguez-Valera
New Abundant Microbial Groups in Aquatic Hypersaline Environments Scientific
Reports 1, Article number: 135 Received 22 August 2011 Accepted 10
October 2011 Published 31 October 2011
Genomic Analysis of Deeply Branching Thermophile Provides Clues to
Early Life on Earth
An Ancient Pathway Combining Carbon Dioxide Fixation with the Generation
and Utilization of a Sodium Ion Gradient for ATP Synthesis
Autotrophic Carbon Fixation in Archaea
©1994-2012 Foster P. Carr
MD all rights reserved
|
