1refers to potentially complete, autonomous element 2motif in Hermes Tpase 3motif in Drosophila P element Tpase 4RVE integrase-like 5REP-Helicase
I Virogenesis/Genesis of Mobile Genetic Elements
Viruses, Plasmids, Transposons, Insertion Sequences, Diversity Generating Retroelements (ref), Retrons (ref), Group II Introns, Group I Introns, IStrons, Integrons, and Transpovirons
retrons, DGRs, CRISPRS, transposons, operons, metagenomes, and archaic viruses
II Mobile Genetic Element Evolution, Content, and Structure
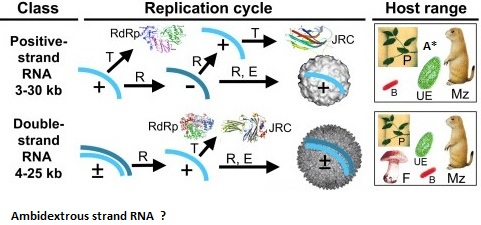
Previous viral classifications have been based on their DNA /RNA types, host cell range, or replicative mechanisms. However new models of grouping recognize architectual, protein structural, and genetic content similarities.
I RNA Ribozymes and Group I Introns arose from the RNA World
II RNA viruses (single stranded and double stranded )and transposition agent developed from the Precellular world and evolved in the cellular world through recombinationsI. Picorna like Group ( Picornavirata) Includes Bymoviruses, comoviruses, nepoviruses, potyviruses, sobemoviruses and (beet western yellows virus and potato leafroll virus)a. Picorna Groupb. Nido Groupc. Levi bacteriophages and Group 2 intron group (Leviviridae of bacteria are ancestral to Narnaviridae of Fungi with evidence of gene loss)d. Astoviruses and Sobemo GroupII. flavi like group (Flavivirata) Tombusviridae, flaviviridae (hepatitis C , pestiviruses), (barley yellow dwarf virus) , and Archael Positive Strand RNA 5.6 Kb (capsid protein similar to nodaviruses, tetraviruses, and birnaviruses)
III. the alpha like group (Rubivirata). Alphaviruses, carlaviruses, furoviruses, hordeiviruses, potexviruses, rubiviruses, tobraviruses, tricornaviruses, tymoviruses, apple chlorotic leaf spot virus, beet yellows virus and hepatitis E virusIV Nodavirus Group
V Recombinant Viruses - Ourmiaviruses (dsRNA + ssRNA = ssRNA replicon and dsRNA Capsid genes)
Double Stranded RNA Viruses -have relationships with the above positive singe strand RNA viruses. Generally classified as Totiviridae- Monopartite , Partitiviridae- Bipartite and Chrysoviridae - Quadripartite Groups. Closer inspection suggest horizontal gene transfers and combinations of the Monpartite groups into the Bipartite and Quadripartite Families.
Familys Birnaviridae , Chrysoviridae, Cystoviridae, Endornaviridae, Hypoviridae , Megabirnaviridae , Partitiviridae , Picobirnaviridae , Reoviridae - includes Rotavirus , Totiviridae, Unassigned genera ,Endornavirus, Varicosavirus
Negative Strand RNA Viruses Order Mononegoviralis originated in the phytopathogenic ascomycete Fungi (>300 Ma)or transfered from plants to these fungi (< 5Ma)
invertebrates
Order Mononegavirales
Family Bornaviridae - Borna disease virus
Family Filoviridae - includes Ebola virus, Marburg virus
Family Paramyxoviridae - includes Measles virus, Mumps virus, Nipah virus, Hendra virus
Family Rhabdoviridae - includes Rabies virus
Unassigned families:
Family Arenaviridae - includes Lassa virus
Family Bunyaviridae - includes Hantavirus, Crimean-Congo hemorrhagic fever
Family Ophioviridae
Family Orthomyxoviridae - includes Influenza viruses
Unassigned genera:
Genus Deltavirus
Genus Emaravirus
Genus Nyavirus includes Nyamanini and Midway viruses
Genus Tenuivirus
Unassigned species:
Orchid fleck virus
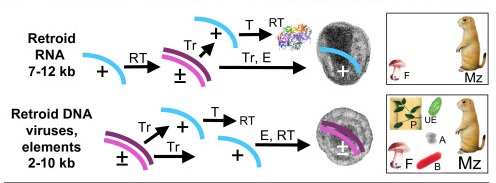
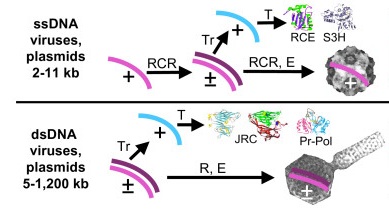
Taastrup virusIV DNA viruses, transposons, and plasmids Evolved from Pre-LUCA, Post LUCA, Pre-LECA, and Post-LECA Worlds
Family origin eco function External Viral form ssDNA/dsDNA
circular/linear Genome Size Capsid Morphology Host. MCP fold PDB Spiraviridae ACV pre-LECA ?pre-LUCA +/- +/- 24893 nt helical rod A-Cren Nanoviridae relatd to RCRE containing elelements +/- +/- icosahedral E-algae/plants jelly roll Gemniviridae ssDNA bacterial plasmid recombinant with +ssRNA Tombusavirus +/- +/- icosahedral E-plants jelly roll Circoviridae Nano + Picorno recominant ambi +/- +/- 1759–1768 nt icosahedral E-mammals jelly roll Parvoviridae releated to RCRE elements +/- -/+ 4-6K icosahedral E-Vert jelly roll Bacillariodnaviridae +/- +/- icosahedral E jelly roll Anneloviridae +/- +/- - 2-4K icosahedral E jelly roll Bidnaviridae +/- +/- icosahedreal E jelly roll Microviridae ?related to circo,parvo, and gemni +/- +/- 3856-6300 icosahedral B jelly roll Inoviridae ? related to ligamenvirales +/- +/- icosahedral B jelly roll Plasmaviridae +/+ +/- mem-pleomorphic B-Mollicutes ? Pleolipoviridae +/+ +/- mem-pleomorphic A-Eu n/a Ligamen virales Lipothrixviridae
-/+ -/+ mem-flexible filaments A-Cren 4helix bundle 3FBL, 3FBZ Rudiviridae -/+ -/+ non flexible rods A-Cren 4helix bundle 3F2E Bicaudaviridae -/+ +/- spindle with 2 tails A-Cren alpha helical 3FAJ Caudo virales Peduovirinae
Teequatrovirinae
T4 like
T4
44RR
RB43
RB49
KVP40 like
Spounavirinae
SPO1-like
Twort-like
Bcep781, BcepMu, Felix01, HAP1, BZX1, PB1,phiCD119,
phiKZ
Mu
Cyanophages
some related to DNA transposon/phages ie. Mu -/+ -/+ icosahedral-ct A-eu, B HK97-like 1YUE Siphoviridae -/+ -/+ icosahedral-nct A-eu,B HK97-like 10HG Podoviridae -/+ -/+ icosahedral-st B HK97-like 2XYY Herpes virales Alloherpesviridae -/+ -/+ icosahedral E-vert HK97-like Herpesviridae -/+ -/+ mem-icosahedral E-vert HK97-like Malacoherpesviridae -/+ -/+ icosahedral E-vert HK97-like Papillomaviridae related to ssDNA viruses with RCRE and SF3 Helicase -/+ +/- icosahedral E-vert jelly roll 1DZL Polyomaviridae related to ssDNA viruses with RCRE and SF3 Helicase -/+ +/- icosahedral E-vert jelly roll 1SVA NCLDV Asfarviridae evolved from phycodna related ancestor -/+ -/+ icosahedral E-pigs E-ticks
2 jelly roll Iridoviridae -/+ ?/+ icosahedral E-invert and vert
2 jelly roll evolved from iridoviridae -/+ +/- oval E-invert 2 jelly roll Mimiviridae -/+ icosahedral E-amoeba 2 jelly roll 1J5Q Phycodnaviridae -/+ icosahedral E-algae 2 jelly roll Poxviridae ?evolved from asfar + rudi related ancestors -/+ -/+ mem-oval E-vert and invert 2 jelly roll Adenoviridae ?evolved from rudi related ancestor -/+ -/+ 26-45K bp icosahedral E-vert 2 jelly roll 1P30 Corticoviridae -/+ icosahedral B 2 jelly roll 2VVF Tectiviridae -/+ icosahedral B 2 jelly roll 1HX6 STIV -/+ +/- icosahedral A-Cren, Eury 2 jelly roll 2BBD Nudiviruses ? evolved from ascoviridae -/+ +/- E-invert Baculoviridae evolved from nudiviruses -/+ rod E-invert ? Polydnaviridae endosymbiosis of nudiviruses-baculoviridae and/or Ascoviridae obligate symbionts of parasitic wasps -/+ rod/fusiform E-insects ? Ampullaviridae -/+ +/- bottle A-Cren ? Guttaviridae -/+ +/- tear droplet A-Cren ? Fuselloviridae -/+ +/- spindle/pleomorphic A-Cren ? Globuloviridae -/+ +/- mem-spherical A-Cren ? Nimaviridae symbionts of rotifers -/+ +/- 300K bp ovoid with tail E-shrimp ? Clavaviridae -/+ +/- 5278 bp bacilliform A-Cren ? Virophages related to DNA transposons -/+ +/- 18343 bp icosahedral A,B,E
Yutin and Koonin propose a complex monophyletic evolution of NucleoCytoplasmic DNA Viruses from Caudiviridae within LECA. They recently reduced the core set of genes for the ancestor from 47 to 30. Their solution is a hybrid of models 2 and 4 .
I propose an alternative paraphyletic model that explains the presence of Nuclear Location Signals in the terminal proteins of Bacteriophage related to NCLDV's .This model is similar to the Yutin-Koonin model but is proposes that the prebacterial and prearchael compartments consisted of a diversity of Eocytes. These Eocytes contained multiple nuclei and were able to differentiate into the archaea, eubacteria, and eukaryocytes in a similar fashion to how bone marrow stem cells differentiate into red bloods cells, platelets, and white blood cells. Byproducts of their diffentiation and Natural Cataclysmic events produced regressed parasitic and symbiotic previral NCLDV life forms for all three cellular domains. Therefore eukaryotic NCLDVs did not developed directly from prokaryotic DNA viruses although there could be some genetic recombination events between the 3 previral DNA life forms prior to or during host infections. Genetic Regression processes occured at the level of Eocyte to DNA virus transformation but subsequent evolutioin in hosts involved cycles of gene additions, recombinations, and gene losses due to natural selective forces.The Eocytes evolved through gene addition processes. Thus the precursor to the free living viral form had gene addition as its main evolutionary force but the immediate descendants had gene loss.
DNA Transposons are Plasmids/Endogenous Viruses that are able to integrate and propagate in Host Chromosomes. They may become Exogenous Viruses by acquiring Capsid or Nucleoprotein Wrappers. Their origin and propagation may be associated with the origin and expansion of Group II Introns and Insertion Sequences. It is unclear if Insetion Sequences arose before transposons or vice versa.
Annu Rev Genet. Author manuscript; available in PMC 2007 December 28. Annu Rev Genet. 2007; 41: 331–368.
Tc1/mariner IS630 TA 1.2–5.0 17–1100 variable 300–550 DD(30–41)D/E HTH (cro/paired) Kolobok H2CH hAT
Ac Group
Buster Group
nd 8 bp 2.5–5 10–25 YARNG 600–850 D(68)D(324)E2 ZnF (BED) P element nd 7/8 bp 3–11 13–150 CANRG 800–900 D(83)D(2)E(13)D3 ZnF (THAP) Galileo
MuDR/Foldback
IS256 7–10 bp 1.3–7.4 0-sev. Kb variable 450–850 DD(~110)E ZnF (WRKY/GCM1) CACTA nd 2/3 bp 4.5–15 10–54 CMCWR 500–1,200 Nd nd TNPA (DNA-binding protein) PiggyBac IS1380 TTAA 2.3–6.3 12–19 CCYT 550–700 DDE? nd PIF/Harbinger IS5 TWA 2.3–5.5 15–270 GC-rich 350–550 DD(35–37/47–48)E HTH PIF2p (Myb/SANT domain) Merlin IS1016 8/9 bp 1.4–3.5 21–462 GGNRM 270–330 DD(36–38)E nd Transib nd 5-bp 3–4 9–60 CACWATG 650–700 DD(206–214)E nd MER37
DD35E Banshee IS481 4/15 bp 3.5 41–950 TGT 300–4004 DD(34)E HTH Helitron (rolling circle)
related to +ssRNA viruses and papilloma/polymova viruses that contain an RCRE
IS91, IS1294, IS801 none 5.5–17 none 5′-TC…CTAR-3′ 1,400–3,0005 HHYY (‘REP motif’) ZnF-like RPA (in Plants) Polintron/Maverick (dna polym)
related to Virophage-Mavirus
none 5/6 bp 15–25 150–700 simple repeat 350–4504 DD(33–35)E ZnF (HHCC) 4–10 DNA virus-like proteins Mutator
IS256? Ginger (related to maverick int?)
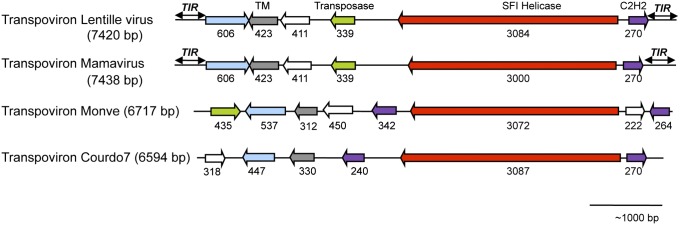
IS3,630,480, 30 gypsy like ltrs H2CH Zisupton Transpovirons 7.5 kb linear and circular.. integrates into virophages and host chromosomes 6 - 8 ORFS
Plasmids, Conjugative Plasmids, ICE's , and AICE's
References
Recombination in Eukaryotic Single Stranded DNA Viruses
New dimensions of the virus world discovered through metagenomics.
Expanding networks of RNA virus evolution
International Committe for Classification of Viruses
Biophysical and biochemical properties of an unusual birnavirus pathogenic for rotifers.
Retroids in Archaea: Phylogeny and Lateral Origins
Protein Conservation in Virus Evolution
Microviridae goes temperate: microvirus-related proviruses reside in the genomes of Bacteroidetes. (icosahedral lytic circular ssDNA viruses)
Related haloarchaeal pleomorphic viruses contain different genome types
Widespread Horizontal Gene Transfer from Double-Stranded RNA Viruses to Eukaryotic Nuclear Genomes
© 2012 Foster P. Carr MD all rights reserved